Alex Lee *
Download a PDF version of this article here.
.Introduction
Protein folding, the process by which a linear polypeptide chain folds into a three-dimensional structure, is an eighty-year challenge that has been described as the final frontier of molecular biochemistry. Despite how long it has remained unsolved, it appears that science is accelerating towards the solution with the advent of neural net AI. Researchers are racing to build, refine, and use tools utilizing these advancements into all stages of drug development. A Google team was just awarded a Nobel Prize for advancements in protein folding prediction. This paper proposes a possible future wherein AI-enabled virtual modeling is perfectly able to predict protein folding and protein interactions. At least for antibody prediction, this horizon is fast approaching.
The advance in technology could help resolve current issues with pharmaceutical patenting. Antibodies have a history of functional claiming due to the technical complexities inherent in the science, with the USPTO even creating a carve out specifically to allow functionally claimed antibodies. Even when it became possible to structurally identify antibodies, courts have accepted that it was uniquely difficult and simply not how the art was practiced. However, there has been a recent move away from this view. In Amgen Inc. v. Sanofi., the Supreme Court affirmed the shift towards a heightened “full-scope” enablement standard for genus claims. Courts have taken note and have begun to trend towards invalidating genus patents for insufficient enablement. Yet beyond the protein folding horizon, pharmaceutical patentees will be able to respond to these changes by utilizing AI enabled protein folding prediction tools. There is a risk that these tools are too powerful and allow patentees to tie up too much in return for too little. Courts should be ready to respond to maintain the patent balance.
I. The Protein Folding Challenge
Proteins are large molecules that perform nearly all the work in a cell.1 They serve as structures, catalysts, hormones, enzymes, and building blocks, and help to execute nearly all cell functions alongside other specialized roles such as antibodies, toxins, or sources of luminescence.2 All proteins found in plant and animal life are made up of varied combinations of just twenty common amino acids.3 They are linked together to form a long unbroken one-dimensional amino acid (1DAA) string that then folds into a three-dimensional shape.4 Most purified proteins will spontaneously refold in vitro after being completely unfolded into its 1DAA chain.5 The three-dimensional structure of a protein determines its biochemical properties because the structure and function of a protein are intimately intertwined.6 Therefore, being able to predict how a protein folds equates to predicting its function. The scientific endeavor to develop a method to predict protein structure is known as the protein folding problem. There are four levels of protein organization.7 At the primary level is the chain of amino acids that make up the 1DAA string. At the secondary level, the chain of amino acids folds into a three-dimensional shape. Common shapes include the alpha helix (where the coils up in a corkscrew shape) and the beta sheet (where the chain folds up on itself).8 At the tertiary level, the secondary structures interact with one another and the entire protein shape forms by folding up on itself.9 Finally, at the quaternary level, multiple tertiary structures may interact with one another to form a final protein.10 This level also includes protein-protein-interaction.
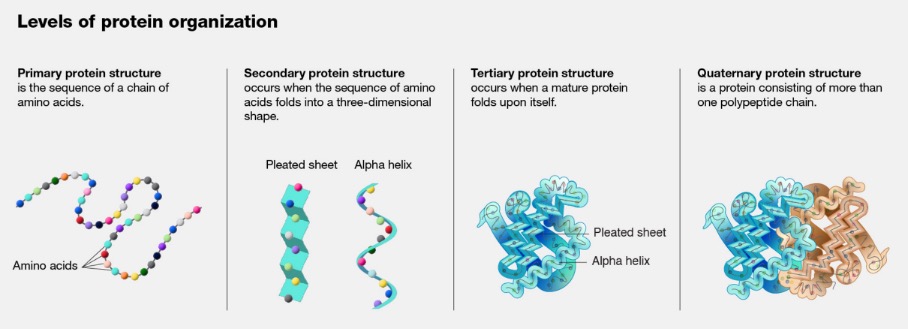
Figure 1. Levels of Protein Organization11
The protein folding problem may be broken down into three questions:12
(i) The physical folding code: How is the structure of a protein determined by the physiochemical properties encoded into its 1DAA chain?
(ii) The folding mechanism: How, despite having an immense number of possible combinations, is a protein able to fold so quickly?
(iii) Predicting protein structure from amino acid sequence: How can a computer algorithm be used to predict a protein’s structure based off of its amino acid sequence?
The accuracy of computer modelling of physical phenomena depends on accounting for all physical forces correctly.13 After a molecule is put in a random initial configuration, the structure is subsequently determined by repeatedly solving laws of physics for the atoms of the protein molecule and the solvent. “Template-based modeling” is when target sequences that are already in the Protein Data Base (PDB) are modeled and tends to be easier.14 By contrast, “free modeling” prediction is when there are no known similar sequences and tends to be more difficult.15 In order to fully utilize this technology, folding prediction must be reliable even when databases are limited.
One major experimental endeavor to study the kinetics of protein folding involved finding folding intermediaries, which are partially structured states along the folding pathway.16 Biochemical pathways almost always have been solved by isolating pathway intermediates and studying their structures. However, this approach has failed with protein folding pathways. Protein folding intermediates exist for an exceedingly short period of time (<1 s); thus one cannot isolate and study them using traditional structural methods.17 This led to the development of new investigatory methods including mutational studies, hydrogen exchange, fluorescence labeling, laser temperature jumps, and single molecule methods.
Despite the myriads of efforts, a complete “folding mechanism” remains elusive. Ken A. Dill and Justin L. MacCallum define a folding mechanism as “a narrative that explains how the time evolution of a protein’s folding to its native state derives from its amino acid sequence and solution conditions.”18 In other words, it is a general principle applicable to a broad range of proteins that accounts for the differences and similarities in folding routes for various proteins.
Researchers have been able to confirm some conclusions. Proteins appear to fold in hierarchical tree structures, rather than linear routes.19 The stability appears to increase as the partial structure develops.20 Proteins also appear to first develop local structures before folding into global structures.21 Scientists are working to unravel the mystery of protein folding, but there is seemingly no shortage of questions to answer.22
We know far more sequences than structures, due to developments in high-throughput sequencing outpacing developments in structure prediction.23 Google’s Deepmind team recently developed a deep learning-based AI tool, AlphaFold2 (AF2).24 While it was heralded as groundbreaking, it still had a substantial error rate.25 Molecular modeling still requires physical, experimental validation. It is unclear when, if ever, modeling technology will reach a point where physical validation of modeling is never required.
A. How Close Are We?
However far we are from crossing the “protein folding horizon” where protein structures and interactions are modellable with certainty, one thing is certain. Researchers have been making tremendous progress. A deep learning neural network AI framework has burst onto the scene, with experts noting that it frequently achieved “an accuracy comparable to that of experimentally derived models.”26 This section details recent improvements in the field and reasons to be believe that the horizon is fast approaching.
1. Community Cooperation
Protein folding is such a grand challenge that organized communal effort is commonplace. Critical Assessment of Protein Structure Prediction (CASP) was one of the first community-wide competitions that spurred advancement, though other competitions have subsequently arisen.27 CASP is held every two years, and each time many different “target sequences” (proteins with structures known only to the organizers) are given to research groups, who test their algorithmic schemes to predict the 3D structures of said targets. Competition organizers then evaluate group performance and publish a paper detailing the results.28 Afterwards, competitors publish their results and methods, allowing the community to learn and improve upon successful methods. With a focus on scientific progress rather than commercial profit, these competitions are important so that advances are immediately disseminated and incorporated into future efforts.
AF2 won CASP14.29 The organizers of CASP14 heralded AF2 as the solution to the protein structure prediction problem.30 The organizers noted that complex deep learning approaches were the most successful. Though AF2 showed substantially improved results compared to its predecessor AlphaFold (AF) in CASP13, the second-ranked model in CASP14 also outperformed AF, showing that groups improved upon AF.31 The success of AF2 continued to expand during CASP15, despite AF2 not even participating.32 AF2 has been open source since 2021, and participants have integrated the AI system into their own approaches, with moderate improvements in accuracy and strides in predicting protein interactions.33 Systems building on AF2 are “approaching the accuracy of experimental methods.”34
There is much anticipation as the impacts of AF2 reverberate throughout the scientific community. After the source code of the software was released, many research papers have cited and utilized AF2.35 Deepmind announced AlphaFold3 on May 8, 2024, with improved accuracy in predicting protein-ligand docking interactions, protein-nucleic acid interactions, and antibody-antigen prediction accuracy.36 The results “show that high accuracy modelling across biomolecular space is possible within a single unified deep learning framework.”37 The Royal Swedish Academy of Sciences recognized the work of the Google Deepmind team behind AF2 when Demis Hassabis and John M. Jumper were awarded the Nobel Prize in Chemistry 2024 for their stunning breakthrough.38
2. Growth of Protein-Structure Databases
Community cooperation has not been limited to competitions. The PDB was established in 1971 as the central archive for all experimentally determined protein structures. It has steadily grown and has been formally maintained by an international consortium known as the Worldwide Protein Data Bank (wwPDB) as a uniform global resource.39 There are currently more than 230,000 protein structures within the database, with the annual deposition rate rising 500% over 20 years from 2000 to 2020.40
After winning CASP14, the creators of AF2 released the AlphaFold Protein Structure Database (AlphaFold DB) in a partnership with the European Bioinformatics Institute.41 While the protein structures have yet to be fully validated, the AlphaFold DB contains 214,683,839 structures, an astronomical increase from the PDB collection.42 This massive data trove contains the predicted structures of nearly every protein known in science and showcases the potential of protein folding technology.43 The AlphaFold DB has been likened to enabling a “Google search” of protein structures for millions of researchers around the globe.44 As recognition for this monumental advancement in scientific research pours in, its impact will be felt throughout various fields and particularly in pharmaceutical innovation.
B. Drug Design
Advancements in protein prediction technology will have significant impacts throughout the scientific community. One area that will particularly be affected is the development of pharmaceuticals.
1. Discovery
The first step of drug development is discovery, where massive sets of drug candidates are reviewed and narrowed down for further research. Projects can start by screening as many as a million compounds to end up with one or two candidate molecules.45 There are multiple stages of drug discovery, including:46
- Target Identification – Identifying biological targets for drugs to bind to that elicit a desired response when bound.
- Target Validation – Validating identified targets through tools such as animal models, monoclonal antibodies, and chemical genomics.
- Hit Identification & Lead Discovery – Screening for compounds that bind to targets with strategies including high throughput screening (HTS), focused screening, fragment screening, structural aided drug design, virtual screening, physiological screening, and NMR screening. After compounds pass screening, the lead discovery stage is used to screen using in vitro assays to characterize both efficacy and safety.
- Hit-to-Lead – Refining hits to produce more potent and selective compounds through the use of structure-activity relationship (SAR) investigations and HTS assays. The multitude of hits from Hit Identification are studied in parallel, and physiochemical and in vitro properties crucial to drug use are characterized, such as solubility and permeability.
- Lead Optimization – Improving deficiencies in the lead compound while maintaining favorable properties. There are various properties and tests to consider, such as high-dose pharmacology, pharmacodynamic (PK/PD) studies, dose linearity, and repeat dosing PK looking for drug-induced metabolism and metabolic profiling.
A variety of screening techniques to identify hit molecules exist. HTS involves screening entire compound libraries against the target using physical assays. This can be an extremely expensive and time-consuming process, requiring the use of laboratory automation.47 This testing assumes no prior knowledge that will aid in narrowing the scope of the search. Focused screening selects from a smaller subset of compounds that are known to likely have some success based on prior knowledge of the protein.48 These strategies have given rise to discovery paradigms using pharmacophores (structural features in a molecule that is recognized at a receptor site and is responsible for the molecule’s biological activity, often shared by compounds that bind to the same target) and molecular modeling to conduct virtual screening.49 Such pharmacophore based virtual screening (PBVS) strategies search large libraries of chemical structures to identify compounds that are likely to bind to the target.50 PBVS techniques will continue to grow more prevalent as libraries of chemical structures expand and modeling programs improve.51 Any of these techniques may be combined with one another; virtual screening can be followed up with in vitro or physiological screening of the compounds.
Companies are already experimenting with virtual drug development. Isomorphic Labs, established under Alphabet, Inc. as a spin off from DeepMind, has partnered with Novartis AG and Eli Lilly & Co. to work on AI-enabled drug discovery.52 A fully AI generated compound developed by Hong Kong-based Insilico Medicine is currently in phase II clinical trials.53 A fully AI generated compound developed by Hong Kong-based Insilico Medicine is currently in phase II clinical trials.
C. Antibodies Beyond The Protein Folding Horizon
Given the importance of proteins to pharmaceutical research, it is easy to imagine a future where near perfectly accurate protein structure and docking prediction is a common tool for research. Such clear insight into the molecular world would revolutionize therapeutic design. Scientists could utilize massively expanded compound data banks that contain information on protein structures and functions predicted down to the atomic level. Scientists may also be able to use AI tools to screen existing compounds, dream up new ones, and refine them. The various physiochemical properties of any known compound would be readily available, and any novel or combined compound would go through reliable simulations to ascertain these properties. The drug discovery process could be done entirely in silico (on a computer), or at the very least with minimal experimental validation. Use of AI is already being integrated into every stage of the drug development process, from drug design and development to designing and running clinical trials.54 Science is accelerating down this path.
A study of FDA approved therapeutic agents between 2009 and 2018 determined that the mean cost of bringing a new drug to market ranged from $314 million to $2.8 billion depending on the therapeutic area.55 Advancements in simulation will likely lower these costs. As protein folding and molecular simulation accuracy improves, drug design methods implementing these new tools will improve in parallel.56 One example is the implementation of AI simulation technology for antibody modeling and interface analysis in drug discovery.57
Antibodies are a type of proteins utilized in certain drugs and treatment methodologies. They are incredibly diverse and connect with complementary large molecules known as antigens.58 It is estimated that an individual has tens of billions of variations in their body at any given time.59 Scientists think that the number of unique antibodies mirror the number of stars in the galaxy.60 These immune receptors have immense biological value as they have the potential to bind to almost any other large molecule, even those that have yet to be conceived.61 Antibodies are valuable because they will only bind with a single antigen, though a single antigen can bind with multiple, slightly varied antibodies.62 This property makes them useful as research tools, therapies, and diagnostics. They are “among the most frequently used tools in basic science research”.63 Antibody based therapeutics are currently the largest class of biotherapeutics, with five of the current top-selling therapeutics being monoclonal antibodies.64 The global antibody market was estimated to be at USD 162.17 billion in 2023 and is estimated to grow at a compound annual growth rate (CAGR) of 11.31% from 2024 to 2032, to be worth USD 425.38 billion by 2032.65
Antibodies are “Y” shaped molecules comprising of two heavy amino acid chains and two light amino acid chains.66 The heavy chain has a variable domain (VH) and three or four constant domains (CH1, CH2, CH3, CH4). The light chain has a variable domain (VL) and a constant domain (CL). Within the VH and VL domains are complementary determining regions (CDR) that vary greatly to determine the antigen the antibody will bind to. The arms of the Y are composed of a light chain paired with the VH and CH1 domains of the heavy chain. This is referred to as the Fab region while the vertical segment of the Y is referred to as the Fc region.
Multiple antibodies may bind to a particular antigen, possibly in different places. The specific place on an antigen that a particular antibody’s Fab region binds to is known as the “epitope.”67 The strength with which a particular antibody binds to a specific epitope is known as the antibody’s “affinity.”68 Finally, how stable the binding is, how long the interaction lasts, is known as an antibody’s “avidity.”69 These various properties of antibodies make them a powerful, customizable tool in scientific research.
Monoclonal antibodies are created using hybridoma technology. Invented by Georges Köhler and César Milstein in 1975, and applied to transgenic humanized mouse strains, hybridoma technology has allowed scientists to generate high quality and fully human monoclonal antibodies.70 The process begins by immunizing a mouse subject with a target antigen, which creates an immune response. Then splenocytes (white blood cells from the spleen) are extracted and fused with immortal myeloma cells, creating a mixture including unfused cells, nonproducing hybridomas, and antibody-producing hybridomas. These are then screened and isolated for a cell line that contains both the antibody-producing ability of splenocytes and the reproducibility of myelomas. This is an immensely resource-intensive step as individual hybridoma cells must be separated and cloned. If successful, a culture of genetically identical hybridomas can be used to produce a single antibody nearly infinitely.
One type of drug likely to be profoundly impacted by the protein-folding revolution is antibody-drug conjugates (ADC), which have been described as “magic bullets.”71 ADCs are complex molecules, combining an antibody, a linker, and a toxin. The antibody acts as a sort of guidance system, to bring the toxin (often called the “payload” or “warhead”) to the site of action. The linker connects the two components and must be stable. ADCs present an opportunity for improved drugs due to the specificity granted by the antibody guidance.72 Unfortunately, due to the complexity of the molecule, ADCs often pose tremendous IP challenges due to the potentially overlapping patent claims from various parties.73
II. Patent Law
To obtain a patent, an applicant must meet four criteria. The patent must be useful,74 novel,75 non-obvious,76 and meet a written description requirement.77 The written description must enable a “person skilled in the art” to make and use the invention in “full, clear, concise, and exact terms.”78 These requirements serve as the statutory screens to protect the “carefully crafted bargain for encouraging the creation and disclosure of new, useful, and nonobvious advances in technology and design” in return for the exclusive monopoly granted by the federal government.79 Though utility patents only offer a limited term of protection, they protect functional features that strongly influence the market success of a product.80 Courts strictly police access to utility patents due to the outsized impact a utility patent can have on the market for a product.
A. AI Challenges
Challenges arise when major changes in the landscape shift the field and pose novel questions to the court of how to apply the rules while staying faithful to the goals of the patent system. The latest such development is the advent of AI. In recent years machine learning AI has made incredible strides with the likes of ChatGPT and Dall-E.81 Though its success is most visible through chatbots and text, image, or video generators, neural net models are disrupting the channels of innovation as well. Legal scholars have begun to grapple with patent law questions that arise with AI. Should AI tools be considered eligible subject matter for patentability, or are they a “basic tool[] of scientific and technological work” that should not be monopolized?82 How should AI-assisted inventorship be treated? So far, the Federal Circuit has held that only natural persons can hold patents,83 but Congress still has room to intervene.84 In addition to posing general questions for patent law, AI development raises specific issues of utility,85 novelty,86 and non-obviousness87 for the pharmaceutical industry. This paper will focus on the enablement and written description requirements for pharmaceutical patents and how they may change with AI-enabled protein structure prediction methods.
B. A Background on Genus Claiming and Enablement
The utility patent system is one that gives nominally uniform rights across various industries, to the benefit of some and the detriment of others. The pharmaceutical industry is one where patent protection is seen to be crucial for innovation.88 In addition to obtaining protection for specific compounds that have been discovered, patentees routinely seek genus claims, which protect “a group of compounds closely related both in structure and in properties.”89 Some scholars argue that genus claims are critical for meaningful patent protection of chemical compounds, as there is a risk that infringers who capture the heart of the invention could avoid liability “by a minor modification of the particular embodiments disclosed in the patent’s specification.”90 If granted, genus claims also afford patentees a broad scope of protection without having to actually make each species covered by the claim. A common technique for genus claiming in chemistry and biotechnology is to draw a core chemical structure with an array of variables around it, representing various chemical groups.91 This claiming practice allows for a large number of permutations within the scope of the claim.
Historically, courts accepted these genus claims, with the CCPA reversing a USPTO enablement rejection on the ground that a more detailed disclosure would force an inventor to carry out an immense number of experiments and discourage them from filing applications.92 The focus was on whether the inventor demonstrated that some species functioned as intended and provided direction for how to test the rest.93 As long as gaps in disclosure could be readily filled by the PHOSITA’s (person having ordinary skill in the art) knowledge, courts would allow broad genus claiming. Particularly in the field of biotechnology, the Federal Circuit upheld genus claims against §112(a) challenges up until the 1990s.94
Antibodies were historically patented through functional claiming, a form of genus claiming. Under this approach, an applicant could claim an entire genus of antibodies by claiming the specific target they bound to or the functions they performed.95 This was due to the technical challenge of structurally describing a complex molecule like an antibody. Such an endeavor was not how the science was practiced. Rather than building towards a molecular structure, hybridoma technology screened samples through trial and error for a cell line that produces the desired monoclonal antibody. Describing every single species was nearly impossible. In recognition of this difficulty, the USPTO has allowed inventors to deposit complex biological materials in a public depository to supplement written disclosure and demonstrate possession of the invention.96 Despite this option, patentees preferred instead to characterize and claim antibodies by their function. For example, this was the case in Noelle v. Lederman, where claims were made to a genus of antibodies simply by characterizing the antigen to which they bound.97
This practice was tolerated by the courts in part due to the complex nature of the science. But there are issues with functional genus claiming.98 Such a claim will include every antibody that binds to a particular epitope or antigen, which might broadly cover over a million molecules.99 This raises anticipation concerns. If a single species out of the million antibodies within the genus claim is previously known, the claim would violate novelty.100 A patentee could attempt to avoid these issues by narrowing the functional claim, such as by claiming an antibody by the epitope it bound to instead of the antigen.101
In 1999, the USPTO acknowledged the technical challenges of characterizing antibodies and carved out a specific exception for antibodies from the usual rule.102 The exception stated that “[a]n applicant may also show that an invention is complete by disclosure of sufficiently detailed relevant identifying characteristics which provide evidence that applicant was in possession of the claimed invention, i.e., complete or partial structure, other physical and/or chemical properties, functional characteristics when coupled with a known or disclosed correlation between function and structure, or some combination of such characteristics.”103 Some offered functional characteristics included “a sequence, structure, binding affinity, binding specificity, molecular weight, and length.”104 All of these allowances recognized the fact that defining antibodies by their underlying structure or genetic sequencing was simply not practical before high-throughput genetic sequencing methods became routine.105
Patentees did not know precisely how antibodies worked, only that they did, posing an enablement problem.106 In Hybritech Inc. v. Monoclonal Antibodies, Inc., though the patentee taught others how to identify, make, and use the claimed antibodies, the trial court found insufficient enablement and invalidated the patent.107 The Court of Appeals for the Federal Circuit reversed.108 The court held that it would be unreasonable to demand perfectly precise calculations of characteristics such as affinity.109 Instead, the court focused on whether “the claims, read in light of the specification, reasonably apprise those skilled in the art and are as precise as the subject matter permits.”110 In the view of the Federal Circuit, the subject matter did not allow for exact precision and therefore the relevant inquiry was whether undue experimentation was required for one skilled in the art to make and use the claimed invention.111 This focus on whether “undue” experimentation was required to “make and use” the invention was affirmed in In re Wands.112
Years later, even after techniques to uncover the genetic sequence or structures were proven, patent applicants were reluctant to claim antibodies by structure.113 Such a narrow claim to a single antibody would be easy to design around, as minor changes that preserved function could enable a competitor to copy the invention while avoiding infringement. This narrow protection would be a worse outcome for the patentee than a trade-secret protection route, due to the enabling specification that teaches the original antibody. Some patentees attempted to craft specific claims while covering trivial variations by including homology percentages in their claims, which set a percentage of similarity to the original claimed sequence that would still be covered.114 However, the USPTO required that claims to homologous groups of proteins would need to disclose the degree of acceptable sequence variations specifically, so as to delineate the metes and bounds of the claim in terms of structure.115 This posed a challenge, as changing even one sequence of an antibody might result in a nonfunctional species, risking rejection.116 Functional claiming of antibodies remained the preferred method until recently.
Recently, the exceptional treatment of allowing functional antibody claims has ended. With its decision in Amgen Inc. v. Sanofi, the Court unanimously affirmed its distaste for functional claiming of antibodies under the enablement requirement.117 The question to ask now is how actors buffeted by this change in wind can be aided by increasing modeling capabilities to meet the higher enablement and written description standards, and how those standards might change in response to the advances.
C. Current State of Written Description
A patent’s specification “shall contain a written description of the invention.”118 In Ariad Pharm., Inc. v. Eli Lilly & Co., the Federal Circuit stated that the distinctive characteristic of description is disclosure.119 A specification is adequately descriptive when it “reasonably conveys to those skilled in the art that the inventor had possession of the claimed subject matter as of the filing date.”120 For the most part, adequate description has two major elements: the enablement requirement and the written description requirement.121 The enablement requirement ensures the patentee satisfies their obligation to disclose technical knowledge in exchange for being granted a patent, so the public may practice the invention.122 The written description requirement forces the patentee to demonstrate that she was actually in possession of the invention that is being claimed at the time of filing the patent application.123
Whether a written description satisfies the requirements varies with the nature and scope of the invention as well as the extent of the scientific and technological knowledge at the time of the invention.124 The inquiry is a question of fact.125 Over the years, courts have developed industry specific standards for enablement.126 On one hand there are “predictable” arts like electrical and mechanical engineering, which require less disclosure as they are rooted in “well defined, predictable factors.”127 Because it is predictable what will occur when circuits are combined, or how much thermodynamic power a newly designed engine will produce, courts have been comfortable with accepting a single embodiment to enable a broad claim.128 On the other hand, inventions in more “unpredictable” arts such as organic chemistry will require more specific and detailed disclosures to avoid the risk of forcing undue experimentation.129
While the USPTO appears to continue to grant broad chemical genus claims as a matter of course,130 federal courts have been increasingly hostile to genus claims under 35 U.S.C. §112(a) for failure to enable or describe the full scope of the claimed invention, particularly for biotechnology and chemistry patents.131 The Court of Appeals for the Federal Circuit has invalidated genus patents by pointing to inadequate guidance in a specification to translate across the full scope of a genus; an excessive amount of experimentation required to parse through the genus; and the lack of precise structural information in the specification to limit the metes and bounds of the genus.132 Several of these decisions have resulted in jury verdicts of over a billion dollars being overturned.133 The trend has been that biotechnology, chemical, and pharmaceutical genus claims lose in court.134 Legal scholars have labeled this the “full-scope” enablement standard and view it as reflecting a shift from a practical focus on whether the disclosure enables others to make and use the claimed invention, to a fruitless endeavor for the exact boundaries of the invention.135 The fear is that functional genus claims are essential for pharmaceutical patent protection—and this new “full-scope” enablement standard effectively kills genus claim patents and guts the protection that the pharmaceutical industry has become reliant upon.136
D. Amgen Inc. v. Sanofi and Genus Claiming
In Amgen Inc. v. Sanofi, the Court took up the enablement question of the degree to which a patentee must show exactly which species within a genus will work as intended.137 The claims at issue involved antibodies that lower LDL cholesterol.138 Amgen owned the 8,829,165 and 8,859,741 patents which claimed all antibodies that bind to the PCSK9 protein and thus lower LDL levels by blocking PCSK9 from binding to the LDL receptors.139 After Amgen sued Sanofi for patent infringement, the District Court for the District of Delaware granted Sanofi judgement as a matter of law, finding Amgen’s claims invalid for lack of enablement.140 On appeal, the Federal Circuit affirmed, signaling their support for the “full-scope” enablement standard by holding that the patents required undue experimentation to obtain antibodies fully within the scope of the claims.141
In order to determine whether “undue” experimentation was required for a PHOSITA to practice the invention, the Federal Circuit applied the following Wands factors:142
(1) The quantity of experimentation necessary
(2) The amount of direction or guidance presented
(3) The presence or absence of working examples
(4) The nature of the invention
(5) The state of the prior art
(6) The relevant skill of those in the art
(7) The predictability or unpredictability of the art, and
(8) The breadth of the claims
While Amgen argued that the no undue experimentation was required due to the embodiments disclosed being sufficiently structurally representative for fulfilling the written description requirement, Sanofi claimed there were millions of potential antibodies that might fall within the genus and require undue experimentation.143 Sanofi pointed to the lack of guidance, the unpredictability of antibody generation, and the substantial degree of trial and error that would be required. The court sided with Sanofi, focusing on the large number of possible candidates within the scope of the claims and the lack of guidance to narrow the field that necessitated a large quantity of experimentation. In discussing the unpredictability of the field of science, the court noted that translating an antibody’s amino acid sequence into a three-dimensional structure is still not possible, and that a substitution within the sequence can alter the function.144 Thus, seemingly the only way to discover undisclosed but claimed embodiments would be through substantial and expensive trial and error.
The Supreme Court granted certiorari to address the question whether Amgen’s ‘165 and ‘741 patents satisfied the enablement requirement of 35 U.S.C. §112(a), such that the invention was described “in such full, clear, concise, and exact terms as to enable any person skilled in the art . . . to make and use the [invention].”145 Writing for a unanimous Supreme Court, Justice Gorsuch affirmed the Federal Circuit’s “full-scope” standard.146 A specification may call for “a reasonable amount of experimentation to make and use a patented invention. What is reasonable in any case will depend on the nature of the invention and the underlying art.”147 What is not allowed is a claim that monopolizes an entire class of antibodies by function simply by disclosing twenty-six antibodies by their amino acid sequences.148
The Court’s opinion cited O’Reilly v. Morse, The Incandescent Lamp Patent and Holland Furniture Co. v. Perkins Glue Co. as examples of prior enablement jurisprudence where overbroad claims were paired with insufficient disclosure.149 In Morse, the claim was too broad and covered all means of telegraphic communication, while the specification did not describe how to make or use them all.150 In Incandescent Lamp, the patentees only possessed an incandescing conductor made of carbonized paper, yet tried to claim “every fibrous and textile material.”151 Such a claim might have been permissible if there was disclosure of a “quality common” to the claimed fibrous and textile substances that made them “peculiarly” adapted to incandescent lighting.152 In Holland Furniture, the claim was to a starch glue and the specification described a key ingredient in terms of its “use or function” rather than its “physical characteristics or chemical properties.”153 The Court took issue with the fact that “elaborate experimentation” was required from one attempting to use the discovery as claimed and described functionally.154
The Court focused on the extreme breadth of Amgen’s claims. When a patent “claims an entire class of processes, machines, manufactures, or compositions of matter, the patent’s specification must enable a person skilled in the art to make and use the entire class. In other words, the specification must enable the full scope of the invention as defined by its claims. The more one claims, the more one must enable.”155 The Supreme Court has affirmed the Federal Circuit’s shift of the enablement inquiry from a question of whether making and using the invention will require undue experimentation to whether defining the full scope of the invention, by experimenting with potentially every species within the genus, will require undue experimentation. The intent seems to be to limit genus claims to species that work, or at the very least limit the genus such that it would not take a prohibitively long time to test every single species within the genus. The Court has signaled a desire for the patentees to do the work to narrow down the genus with some principle before applying for a patent, rather than seeking to claim potentially millions of compounds based on function.
Though this full scope view of enablement has prevailed, USPTO guidelines post-Amgen state that the Wands factors still control.156 The view of the USPTO is that enablement turns on the degree of experimentation required by the specification and whether it is “reasonable.”157 Yet in Baxalta Inc. v. Genentech, Inc., the Federal Circuit relied on the “full scope” test from Amgen to affirm a district court decision of invalidity for lack of enablement.158 Baxalta’s patent covered millions of antibodies, while the specification disclosed just eleven amino acid sequences. Like in Amgen, the court took issue with the fact that nothing in the specification taught a PHOSITA how to identify antibodies that fell within the claim limitations other than by repeating a brute force trial-and-error process.159 There was no way for a PHOSITA to “predict” which antibodies would perform the claimed functions.
With Amgen Inc. v. Sanofi, the Supreme Court has backed a strict, higher bar for enablement, especially for functional genus claims pertaining to antibodies where potentially millions of compounds are claimed with minimal disclosure. When a novel drug target is discovered, it is likely a naturally occurring phenomenon that cannot be patented, as it already exists inside the body of a person.160 Discovering a novel target that elicits a desired pharmacological effect is analogous to discovering properties of electricity in Morse,161 or BRCA genes in Myriad,162 or the pre-modified bacteria in Chakrabarty,163 all of which were unpatentable. Discovering such natural phenomena is a valuable contribution to society, but granting a broad patent to all applications of that discovery would “shut the door” on future innovation and inventions that may improve upon the initial disclosure.164 Such a broad claim without similarly broad disclosure is impermissible. What may be patented is a specific application of said natural phenomenon, like the twenty-six antibodies disclosed in the specifications in Amgen, or the telegraph as one method of electromagnetic communication in Morse. The unpredictable nature of the science and protection against minor variations warrant some broader scope of protection than the four corners of the specification, but this is countervailed by the interests against precluding the field from future improvement and the reciprocity of the patent bargain. With these policy underpinnings in mind, the next part examines how advancement in predictive simulation might affect claiming and enablement.
III. Folded Enablement
Thus, the Court has signaled its dissatisfaction with broad functional genus claims particularly pertaining to antibodies that offer little to no instruction for a PHOSITA to narrow the genus. Parties working in drug development may feel limited to specific compounds in their claims and therefore vulnerable to minor variations. Advancements in simulation technology utilizing AI may offer a solution. Though functional claiming of antibodies is effectively voided, increased modeling capabilities will enable scientists to both predict the structures of the compounds claimed and hypothesize a genus of molecules that have similar functions.
These advancements may directly address the concerns raised by the Court relating to functional genus claiming of antibodies. In Amgen, the Court noted that antibody science was unpredictable and that scientists cannot “always accurately predict exactly how trading one amino acid for another will affect an antibody’s structure and function.”165 It also took issue with the lack of guidance given to narrow the large breadth of the claims.166 When an actor designs a drug around a novel target beyond the protein folding horizon, they will be able to not only disclose the specific structures of the species that they synthesized and tested, but also have ways to narrow down the potential genus of compounds that bind to that target. They also may be able to simply disclose hundreds of species in their specification. In silico simulation can be used to screen and filter for prior disclosures to avoid anticipation, for nonfunctional species to avoid enablement issues, for any other characteristics that might make a species an unattractive candidate for drug development, and even to generate a proposed procedure to synthesize the remaining species. The question of nonobviousness may be looming, but application of this technology towards a novel target may be enough for courts.
The resulting patent application might look like a specific disclosure of twenty-six antibodies that were produced and tested alongside a genus claim consisting of merely 300 species that have been simulated to be found as viable but not preferrable alternatives. Or it might look like a specific disclosure of 326 antibodies that were simulated to be the best candidates to bind to a particular target, with experimental validation on twenty-six of the group. The characteristics evaluated by the program to narrow down the genus could be fully disclosed as guidance as per the second Wands factor.
Both hypothetical claims would likely meet the full scope enablement standard that is the law following Amgen, even though the Wands factors will take into account the advances in simulative capabilities and the science. Courts will recognize that the skill of those in the art has risen, that the art has become more predictable, and consider the state of the prior art in considering whether undue experimentation will be required. Narrowing the claimed genus would address the major concern in Amgen and help the court find that undue experimentation is not required. To get around anticipation or obviousness objections, patentees may structure their claims as applications of these compounds as a means of binding to newly discovered targets.
A. Claim Complexity
Rather than enabling patentees to claim compounds structurally, AI-enabled advancements in simulation technology may have the opposite result. If the technical capability to filter a broad library of compounds based on functional criteria exists, it may also sufficiently enable a PHOSITA to define the full scope of a functionally claimed genus without undue experimentation. Courts may be willing to accept functional claiming of an antibody genus by a patentee that provides sufficient narrowing criteria and disclaims nonfunctional species. The question would be whether the burden of sorting through an expansive genus to clarify the bounds of a claim should be placed on the patentee or on the public.
An inventor has no incentive to disclose any more than is required by law.167 Any disclosure that is not claimed is dedicated to the public.168 It may be that a heightened burden on the inventor will increase the costs of innovation. This may slow the pace of invention and dissemination of knowledge. However, there are benefits to imposing the burden on the patentee to clarify the scope of a claim. In a competitive industry like pharmaceuticals, actors will be both patentees and patent readers. Having clear patent claims will allow readers to design around the patent to improve upon the inventions of their competitors.169 This would stimulate competition, an integral goal of the patent system. If a competitor is left to sift through a functionally claimed genus, they may be met with uncertainty when deciding whether to utilize a compound at the outer fringes of the claim. This leaves the competitor with three unfavorable options. They may try to negotiate a license, which leaves them at the mercy of the patentee. They may simply choose to abandon the attempt, which may deprive the public of innovation. Finally, they may attempt to find relief through litigation, such as an action to invalidate the patent or find noninfringement. This is a costly process that should be avoided if possible. While it is important to protect a patentee from infringement by trivial variations, the burden for the practicing public to narrow a functionally claimed genus must not be too great.170
Another potential issue is the increasing complexity of claims. Claiming compounds functionally has the benefit of simplicity. It is possible that the kind of structural claiming enabled by simulation technology greatly raises the information costs of patent readers when deciphering the claim. Readers may require access to certain computer programs. The group having “ordinary skill in the art” may be exceedingly small, limited to those with expertise in computational biology.
Presently, biochemical patent claims are inaccessible to many. While individual inventors may be disproportionately affected by rising information costs than those under the umbrella of large firms with deep pockets, the typical pharmaceutical patentee is not an inventor working out of her garage. The average skill required to understand and make use of a pharmaceutical patent is already high. Claiming compounds structurally may raise the complexity of reading claims, resulting in lowered access for some, but with a net positive effect. The practicing public will have more definite notice of what is claimed by the patent, spurring innovation by allowing competitors to design around the patentee. Parties with the resources to compete in the pharmaceutical market will adapt to reading the more complex claims. In the end, the twin goals of innovation and access will continue to be met.
B. Means Plus Function Plus Simulation
In response to the movement away from functional claiming of antibodies, Professors Mark Lemley and Jacob S. Sherkow proposed a middle ground involving means-plus-function claiming.171 35 U.S.C. §112(f) provides the statutory basis.172 Despite explicitly permitting functional claiming, a §112(f) claim is actually substantially narrowed in scope.173 Such a claim is not construed to cover every means of performing the claimed function, and is instead limited “only to those disclosed in the patent’s specification and equivalents thereof.”174 As applied to antibodies, this serves as an intermediary between broad, purely functional claims and narrow species claims. A claim for means of binding to a target antigen accompanied with a limited specification would not be found invalid for written description or enablement as it would only be construed to cover the exact species that are disclosed in the specification.
The fight then is over what constitutes an “equivalent” and what test should be applied to accused infringers. Warner-Jenkinson Co. v. Hilton Davis Chem. Co. says there is equivalence if the accused and claimed thing perform “substantially the same function in substantially the same way to obtain the same result.”175 The traditional aim is to capture later-developed equivalents that are unknown at the time of applying for the patent. Though unlikely, it is possible that two structurally different antibodies have the same function: they bind to the same epitope, have the same binding affinity, have the same avidity, and thus perform substantially the same function in substantially the same way to obtain the same result. It may be a question of the degree of scrutiny that is applied to the measurements of these characteristics to find equivalence.
The species within the disclosure cover literal infringement. There is a separate doctrine of equivalents (DoE) that would apply as well. For means-plus-function claims, this DoE applies in two circumstances: where function is similar but not identical, and where the equivalent did not exist at the time the patent issued.176 For a precise science such as antibodies, the first application would not apply. Function would have to be identical to be covered as binding to the same antigen at a different epitope should be sufficiently different. There is the possibility then that means-plus-function equivalence applies DoE onto the equivalent structures covered by literal infringement.177 Lemley and Sherkow argue this strategy can capture structurally different antibodies that share functional characteristics while being sufficiently narrow to pass under the full scope enablement standard. The basis for written description – preventing gun jumping and late claiming – will be supported because structures must be disclosed when filing, so patentees will need to identify and possess the antibodies before claiming.
But the view is different on the other side of the protein folding horizon. Patentees would be able to identify and disclose a significant number of species within the specification. Means-plus-function claiming enabled with sophisticated simulation may wind up with hundreds of disclosed species, and allowing claims to extend to equivalents may wind up crowding out the field for future innovators. It is conceivable that within the hundreds of disclosed species, some may be strategically added not for their value as viable drug development candidates, but to widen the net of equivalents so as to fend of undiscovered improvements. Courts will likely want to narrow what is covered under equivalents by having a high standard for similarity. Compounds will be screened based on characteristics such as binding epitope, binding affinity, avidity, among others. To encourage improvements in a mature field, courts may want to rule that in order for a compound to infringe on an equivalent, it must match every characteristic to an extraordinarily high degree.
C. How Much is Enough?
Advanced molecular simulation will enable actors to run extensive screenings and simulations of compounds at minimal costs. This raises the question of how much patentees should be expected to do in order to claim a compound. Would it be proper to allow a claim to a genus of 10,000 antibodies as long as a computer has generated a “recipe” to synthesize each one, and simulated the interactions with a target epitope with a positive outcome? Should even a 99.9% confidence in in silico reliability be enough? While requiring some degree of physical validation may serve to ensure that the public is not swindled out of a proper patent bargain, could such a requirement ultimately be an undue burden on inventors that will ultimately stifle innovation?
The labor in creating a list of compounds may be relatively trivial. The scientific endeavor to advance protein folding simulation is a worldwide communal effort and the fruits of that endeavor should be a commons to be enjoyed by all. While there may be valuable work in discovering a novel target, the work to screen existing databases generated through common effort is not proportionate to a monopoly on 10,000 antibodies for use in binding to a specific epitope. Such a grant might fence off the field and block others when only a select few antibodies will be used in clinical trials and ultimately only one may possibly make it into a drug. Within the 9,999 other antibodies that were screened and deemed to be inferior choices, there may be one that is superior in different circumstances such as use at high altitudes, or on women.178 Or there may simply be one that was erroneously predicted to have an unfavorable characteristic but in reality, is superior. Precluding others from experimenting with the remainder of the genus may deprive society of valuable advancements.
With that being said, an inventor deserves to have exclusivity and protection for their invested efforts. That protection should account for trivial variations so that infringers cannot avoid liability with minimal effort. Without the guarantee of protection against follow-on actors, few will want to undertake the costly process of developing a pharmaceutical compound and obtaining FDA approval to bring it to market. Adding a requirement that a patentee not only successfully synthesize the full genus that they claim but validate that each species functions as predicted might be a prohibitively expensive and tip the scales on the cost-benefit of innovation. As future courts are faced with these difficult questions, they should maintain the balance between twin goals of innovation and access. Innovators must be protected so that they continue to have an incentive to progress science and the useful arts. The bounds of patent claims must be clear so that the public has proper access to improve upon the invention.
Conclusion
Protein folding and related technologies are developing at an exponential rate. We may soon cross the horizon into a world where the molecular mysteries of complex proteins are made clear for researchers. Despite the history of functional claiming of antibodies and genus claiming, courts have transitioned into a full scope view of enablement after Amgen Inc. v. Sanofi. This status quo may make it difficult for pharmaceutical actors to obtain protection against minor variations of their inventions. Advancements in protein folding and interaction may offer the solution. Modeling capabilities will enable actors to structurally define not only the compounds that they are using in physical trials, but also any functional equivalents within the entire theoretical universe of molecules. This will allow patentees to meet the full scope standard enough to protect against variations and equivalents. There is a risk that this trivializes the enablement standard and allows inventors to tie up too much in return for too little.
